Artificial intelligence (AI) has become a promising tool for the discovery of pharmaceuticals, as it can enhance the probabilities of success and adds greater precision, speed and profitability to a process with high rates of failure, extensive development periods and high costs.
On this occasion, we will discuss a process known as virtual screening. This refers to digital solutions that are developed to identify new molecules for specific cellular targets, thus accelerating the discovery of new pharmaceuticals. To date, the lack of available protein structures and the lack of diversity of the collections of compounds has prevented a more generalized use of this approach. Currently, the availability of extensive and diverse virtual chemical libraries and access to the structures of nearly all proteins represent two fundamental advances that make the successful adoption of virtual screening possible. Furthermore, the replacement of classical coupling tools with coupling procedures based on machine learning will unleash the full potential of virtual screening, making the discovery of pharmaceuticals more efficient, profitable and affordable for many companies in the biotechnology sector with modest budgets.
THE PROCESS OF DISCOVERING A NEW PHARMACEUTICAL TREATMENT
The search for a new pharmaceutical treatment begins with the identification of chemical compounds or molecules that can serve as prototypes for medication (‘candidate pharmaceuticals’) intended for a known and relevant therapeutic target for a certain pathology. The first step is high-throughput screening (HTS), an automated testing process in which chemical compounds are tested to discern their biological activity and therefore, therapeutic potential based on the effect shown in this test. The more extensive and diverse that the collection of analyzed compounds is, the greater the possibility of finding a molecule that could become a pharmaceutical. And while HTS is an advanced technique that lowers costs and has drastically enhanced the efficiency of searching for candidate pharmaceuticals, access to collections of large and diverse chemical compounds can be costly and screening millions of molecules could take months.
Genetic Fingerprint by Topazium
VIRTUAL OR IN SILICO SCREENING
Virtual screening is analysis through simulation or modeling that is conducted virtually (‘in silico’) on a particular target therapy with several billions of virtual chemical compounds.
This technology could identify small molecules (ligands) that can bond to a therapeutic target and modify their function, thus correcting the damaging role they play in the development of a particular disease. The ‘in silico’ demonstration of the interaction between a molecule and a biological target does not represent the confirmation of its beneficial biological activity as the compounds identified through virtual screening must be subjected to biological testing in a laboratory to verify and quantify their interaction with the therapeutic targets that were tested. But virtual screening offers a quick and effective way to preselect a subset of molecules for subsequent testing, avoiding the need to analyze entire collections of compounds.
How does it work?
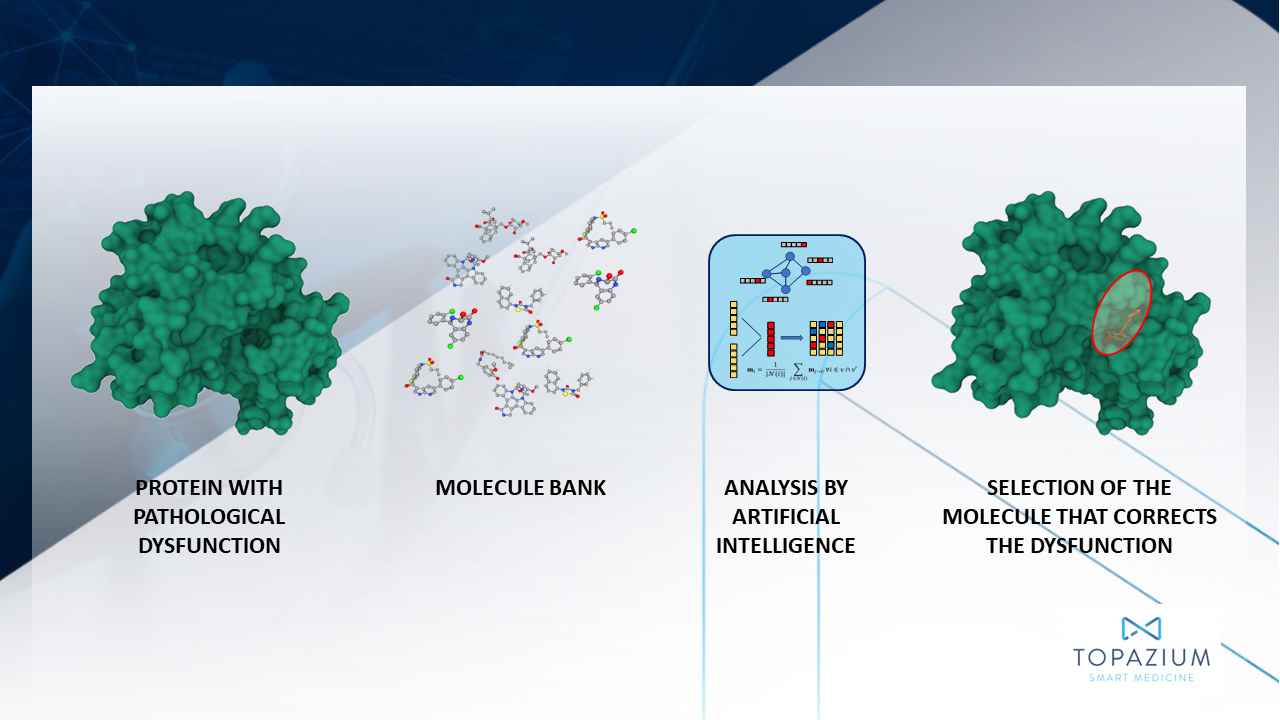
The therapeutic target is a biological molecule that plays a fundamental role in the disease development process on a cellular level. Once the relevant target has been identified, chemical compounds are sought to bond to it and modify its activity, thus correcting the problem. It is important to note that most therapeutic targets that are currently used are proteins. In order for the virtual screening to be successful, we must know as much detail as possible about the three dimensional (3D) structure of the therapeutic target. Traditionally, the precise determination of the 3D structure required sophisticated optical technologies, such as x-ray diffraction or cryo-electron microscopy. However, recent advances in various AI tools like Helixfold and Alphafold have made it possible to predict the 3D structure of nearly all proteins found on living organisms with a high level of precision, making virtual screening a viable option to find chemical compounds that could bond to practically all possible therapeutic targets. While HTS requires access to real chemical compounds, properly stored in vials in specific collections and the ability to synthesize them in the amounts needed for the testing, virtual screening uses virtual collections. Therefore, virtual screening not only gives pharmaceutical discovery the effectiveness and savings that pharmaceutical companies need; it also can greatly improve the likelihood of success by significantly increasing the diversity of the searchable chemical space.
CHALLENGES AND ADVANCES IN VIRTUAL SCREENING
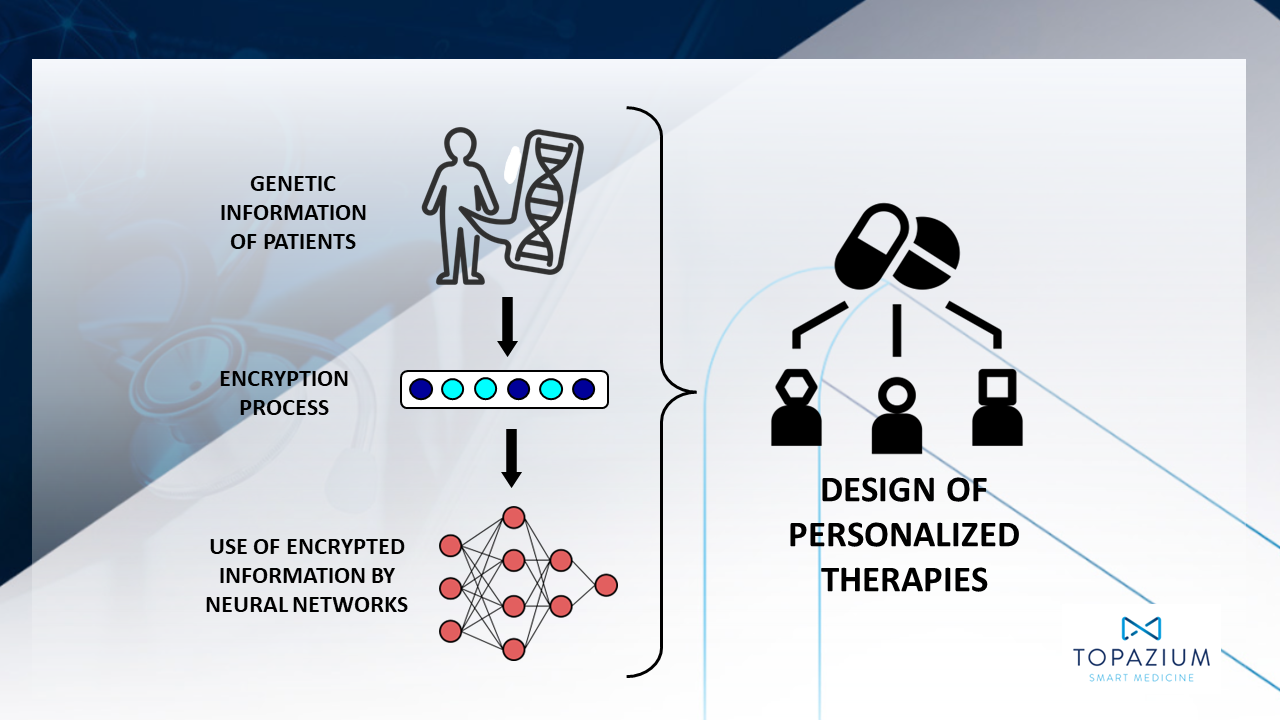
The first major challenge in the strategy to discovery new pharmaceuticals is the identification of a physiologically relevant target. Currently, targets are mainly identified through genome sequencing or genetic expression studies. However, analysis of this type of information is very costly and takes a great deal of time. AI is a highly effective technological response to overcome this challenge. For example, Topazium uses a tool called Genetic Fingerprint that uses genetic information from patients with cancer to identify possible unique therapeutic targets. Using technology similar to what is used to create cryptocurrencies or encrypted information systems, the tool codifies this information in order to generate a ‘virtual tumor genome’ for each patient whose clinical evolution can be studied. Therefore, the tools makes it possible to select those genomes that are associated with a worse prognosis in order to identify the most representative genetic mutations that are responsible for unfavorable clinical development. These harmful mutations reveal intervention points on which pharmaceuticals can act to attack tumor cells, fostering the development of personalized therapies.
Genetic Fingerprint by Topazium
On the other hand, thanks to knowledge of the three-dimensional structure of the target proteins and the access to virtual collections of molecules, researchers can conduct a virtual screening to analyze the affinity with which the molecules are predicted to bond to their target. A computer simulation known as molecular coupling makes it possible to explore different configurations and orientations (collectively called ‘poses’) that a small molecule can adopt within the natural crevices (binding sites) of a protein structure). Through this analysis, researchers look for certain positions that are especially favorable due to the thermodynamic state of the molecule; particularly the level of free energy associated with the position. The molecules that have the most favorable positions are considered ‘hits’ and move on to the next stage of pharmaceutical discovery.
Advances in machine learning (ML) in the past two years have dramatically improved the coupling procedures, mainly by considering the dynamic structures of both proteins and ligands in solution. New ML tools introduce significant improvements in the performance and precision of the coupling procedures. Their use in parallel with high performance computers is expected to represent a giant step forward in terms of the efficiency and reliability of virtual screening, making the most of the rich structural information provided by predictive systems of the 3D structure of proteins and the wealth of chemical diversity offered by the large virtual collections.
In short, AI can currently help us to discover new uses for existing medications (repositioning), identify new candidates for pharmaceuticals and design new chemical compounds that could allow for new treatments for rare or terminal diseases.
Carlos Galmarini
CEO Topazium
Comments on this publication