In 1953, when Francis Crick, James Watson, Maurice Wilkins and Rosalind Franklin unveiled the structure of DNA, one scientific race ended and another began: once it was known that DNA was an ordered sequence combining four bases, adenine (A), thymine (T), cytosine (C) and guanine (G), it was necessary to discover how nature used this information to construct a protein, a chain made up of 20 types of amino acids. The translation of one into another implies the existence of a code, the genetic code. But a code can also serve other purposes, and for years researchers have been trying to use it as a medium for storing digital information that could survive even the extinction of humanity.

It was physicist George Gamow who first proposed that amino acids are encoded by units made up of three bases (triplets). In 1966, the contributions of several scientists deciphered the genetic code, which consists of 64 triplets or codons—four elements (A, T, G and C) in groups of three—and their correspondence with the 20 amino acids. The two numbers are not the same because the code is degenerate (redundant): several codons code for the same amino acid during protein synthesis.
Codes and algorithms for conversion into DNA sequences
The idea of using the genetic code to store other kinds of information soon piqued the curiosity of scientists. In the early 1960s, physicists Richard Feynman and Mikhail Samoilovich Neiman, along with mathematician Norbert Wiener, speculated on this possibility. In 1988, artist Joe Davis and Harvard University implemented the use of DNA as a code in the Microvenus project: the figure of an ancient Germanic rune was represented as a simple map of bits (zeros and ones), which was translated into a DNA sequence by means of a code, and this sequence was inserted into a bacterium.
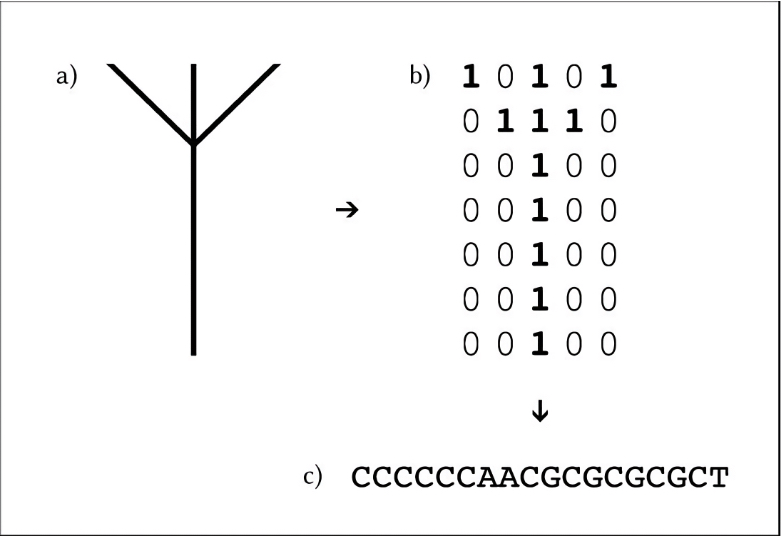
In 2008, biotechnologist and entrepreneur Craig Venter created the first synthetic genome in a bacterium. To differentiate it from the original, he inserted the name of the institute and some of the authors of the paper into the DNA sequence as a sort of watermark—a verification used in transgenic organisms. Venter employed a literal alphabetical code, using the letter designation of each amino acid to write the words; the problem is that there are only 20 amino acids, and the vowels O and U and the consonants B, J, X and Z are missing. In 2010, Venter’s institute unveiled a self-replicating synthetic bacteria watermarked with a web address, quotes from Feynman and the writer James Joyce, and another alluding to the physicist Robert Oppenheimer.

But the purpose of DNA encoding is not to hide secret messages, let alone use three DNA bases for each letter, but to store any digital file in a compressed form. A direct alphabetical translation is therefore of little use. Various codes and algorithms have been designed to convert binary data into DNA sequences and vice versa, introducing redundancy and verification systems to avoid errors, something similar to the check digit in bank account numbers. Texts such as Shakespeare’s sonnets or books by Archimedes and others, but also music albums, images, the entire English Wikipedia or an episode of the Netflix series Biohackers have been translated into DNA. The ill-fated Israeli space probe Beresheet-1, which crashed on the moon in 2019, carried an archive of 20 books and 10,000 images in DNA format.
Artificial fossils of encapsulated DNA
The advantages of DNA over current digital archive storage systems are mainly the density of the information and its durability. In terms of the former, according to Latchesar Ionkov, a computational scientist at Los Alamos National Laboratory working on a project in this field, the estimated 33 zettabytes (billions of gigabytes, GB) of digital information that humanity will generate annually by 2025 would, in DNA form, fit inside a ping pong ball. Another estimate is that one gram of DNA could store 215 petabytes (millions of GB).
As for durability, it will depend on the container of synthetic DNA. One common method is to break the DNA into fragments and introduce them into bacteria that perpetuate the information as they multiply; the downside is that over time there will be mutations, changes in the sequence that can make the information unreadable even with robust verification codes. Faced with this problem, Robert Grass’s group at the Swiss Federal Institute of Technology in Zurich (ETH) has created artificial fossils: silica nanospheres, like grains of sand, containing encapsulated DNA that can be dissolved if the information needs to be recovered, but which can last for 2,000 years at a temperature like that in Zurich, and over two million years if kept at -18°C, for example in the Global Seed Vault in Svalbard.
There are still major technical challenges to overcome. DNA synthesis and sequencing has advanced enormously, but the speed is still slow and the cost high: Ionkov’s project aims to write one terabyte (TB) and read 10 TB within 24 hours for $1,000, which will still be too slow and too expensive. It will be decades before we have DNA recorders and players that integrate all the steps and are fast, cheap and error-minimising. But experts say solutions are urgently needed because the amount of digital information we generate annually is growing faster than the storage capacity installed in data centres. What happens if the day comes when there is not enough storage in the world for all our information?
Comments on this publication