According to many expert bacteriologists, epidemiologists and the odd virologist, if science doesn’t prevent it, in a few years we could return to a stage of pre-antibiotic humanity, i.e. those dark years where bacterial infections were at their worst without an effective weapon to stop them.
From the standpoint of the antibacterial fight, we could always resume that research, which was very advanced in the former USSR, on possible bacteriophages –viruses that infect bacteria, as potential therapeutic agents. While these omens, good or bad, follow their own course, new research on the cellular and molecular mechanisms of antibiotics and the generation of resistance, will always be very well received.
Bacterial resistance
Bacteria generating resistance to antibiotics— that of mutations, selective pressure and evolution— is much more serious than we think, since such resistance can be transmitted from some bacteria to others by mechanisms rather than specific ones. We must also remember that we all have our share of guilt in the generation of these resistances. Taking antibiotics like we’re possessed even when our bunions hurt or not finishing the treatment once started is the breeding ground, never better said, where resistant bacteria find their small Shangri-lá.
A study led by researchers from the Spanish National Research Council (CSIC) has identified a new family of enzymes involved in transferring antibiotic resistance between bacteria. The paper focuses on the role of these enzymes, called relaxases and a DNA fragment that is essential in the genetic exchange between Gram-positive bacteria. Gram-positive bacteria are those with a thicker cell wall. Many are pathogenic, such as those that produce tetanus, botulism or gangrene. The results have been published in the journal PLOS Genetics.
Genes that provide resistance to antibiotics are often found in genetic elements that can be transferred from one bacterium to another. This process of transferring an emitter cell to a receiver is what is known as conjugation. As explained by Wilfried Meijer, researcher of the CBMSO (CSIC-UAM joint center) and work director, relaxase is essential in the first steps of that genetic transfer, as it introduces a cut in one of the DNA strands to be transferred. This allows this DNA to start being copied and a strand to be released which is to be transferred to the receiving cell via a connection channel.
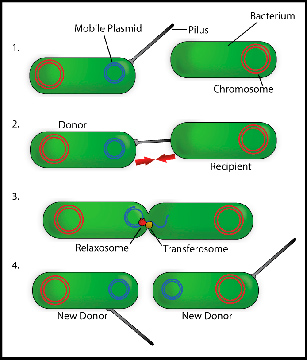
The researchers responsible for this study have been able to identify the gene that codes the relaxase of a conjugative element— the element that is part of the mechanism transferring information from one bacterium to another— used as a Gram-positive bacteria model system. This breakthrough has highlighted more than 800 genes that encode similar relaxases in other bacteria. As mentioned, almost all of these genes are located in Gram-positive bacteria, and a large part of them are present in the intestinal microbiota.
These results represent an important advance because they give rise to studies of conjugation in bacteria of microbioma —the set of bacteria that live with us— and, especially, the knowledge we have about the dissemination of genes.
Check the original text on the website of Fundación madri+d
José Antonio López Guerrero
Tenured professor of Microbiology
Director of Scientific Culture of the CBMSO
Comments on this publication