In 1829, three decades before the publication of The Origin of Species by Charles Darwin, the first Neanderthals fossils were discovered in Belgium—thus they were not suspected of belonging to another extinct human species. It was only when scientists like Thomas Henry Huxley began to apply Darwinian theory to humans that our evolutionary link with other primates began to be drawn.
During the following decades, the discovery of new fossils began to supply evidence of the evolution of the human being from other species. The science of palaeoanthropology was born, dedicated to studying the morphology and making anatomical comparisons between the different fossils to understand what Huxley called “the place of man in nature.”

However, more than a hundred years of science have revealed that the story is extremely more complex than the simple and primitive idea of a “missing link.” Today we know that our family tree is rather a weave of innumerable ancestral species that intersect, a panorama so complex that it has led the renowned paleoanthropologist Meave Leakey to say that we still have another hundred years of research to do before coming to understand the history of our evolution.
The reinvention of the study of human evolution
In the last decades, another scientific discipline has come to the aid of palaeoanthropology, prompting an authentic reinvention of the study of human evolution. In 2010, the existence of an old branch of our family was discovered thanks to an approach that reaches where the anatomical study does not. In this case it was also based on a fragment of bone, but instead of preserving it intact as a valuable relic, the researchers pulverized the fragment to scrutinize its most intimate structure, that of its DNA.
The discovery of the Denisovans, the possible human species or subspecies originally identified through their DNA that lived in Asia until about 40,000 years ago—some studies suggest perhaps 20,000 years ago or even less—was in fact the culmination of a long scientific journey that had been going on for decades. The emergence of molecular research in human evolution began to gain strength in the 1960s thanks to the analysis of proteins. In the 1980s, when many of today’s molecular biology techniques had not yet been invented, scientists such as the Swedish geneticist Svante Pääbo undertook the laborious task of sequencing DNA thousands of years old.

Initially the researchers focused on mitochondrial DNA, smaller than nuclear DNA and present in multiple copies in each cell, making it easier to read. But now, in the 21st century, technological progress has led to an explosion of ancient DNA sequencing that has allowed the analysis of nuclear DNA samples up to hundreds of thousands of years old.
The record for the oldest human DNA
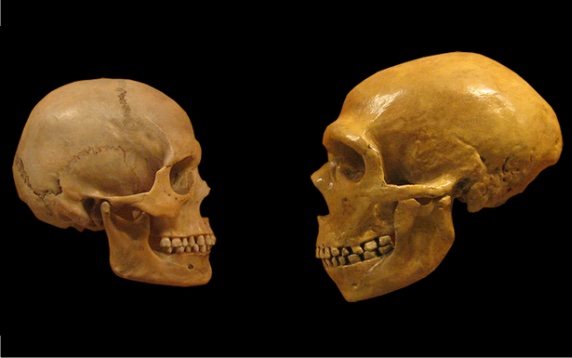
Since 2016, the record for the oldest human DNA has been held by a fragment of the nuclear genome of a Neanderthal ancestor that lived 430,000 years ago in Sima de los Huesos in Atapuerca (Spain). When samples are preserved frozen in permafrost, much older genetic fragments can be recovered; for example, in 2021, DNA was sequenced from the molar of a mammoth that lived 1.2 million years ago. We now know the genomes of Neanderthals and Denisovans, and the various remains found have detailed a complex process of hybridisation between the different species that would have been impossible to unravel using the methods of classical palaeoanthropology. Genomic and epigenomic studies—the latter being chemical modifications of DNA that do not alter its sequence— have even made it possible to assign physical traits to these extinct humans: according to the DNA of a young woman who lived in the Siberian cave of Denisova 75,000 years ago, these individuals were close in appearance to Neanderthals, although with certain differences, such as a broader face and a wider jaw.
Palaeogenetics, or the analysis of ancient DNA from fossil bones, has revolutionised palaeoanthropology, to such an extent that in 2022 the Nobel Committee deemed Pääbo worthy of the Physiology or Medicine prize, the first time in its history that the Swedish awards have distinguished this branch of science, which traditionally did not seem to fit into either category. Progress in this field has been astonishing, and today it is going much further, even dispensing with the need for skeletal remains. In 2003, geneticist Eske Willerslev of the University of Copenhagen (Denmark) began recovering so-called environmental DNA directly from frozen soil and cave sediments. In April 2017, a team led by the Max Planck Institute for Evolutionary Anthropology (Germany) obtained for the first time mitochondrial DNA from Neanderthals and Denisovans from the soil of several caves, up to 130,000 years old.
Beyond the human family, the study of ancient environmental DNA is also providing insights into the animal and plant communities that once inhabited terrestrial regions, in a way that would be impossible with traditional methods. In 2022, environmental DNA was sequenced from Greenland permafrost that dates back 2 million years. It’s the oldest known DNA to date and it reveals an ancient ecosystem in which mastodons, reindeer, hares, rodents and geese lived in mixed forests of poplar, birch and thuja trees.

Paleoanthropologist Antonio Rosas, from the National Museum of Natural Sciences of the Spanish National Research Council, and co-author of the study that recovered environmental DNA from Neanderthals and Denisovans, explained to OpenMind that the work was an exploration of the possibility of recovering DNA without bones in places that have been “lived in, eaten in and defecated in.” A placenta from a birth or a haemorrhage, adds Rosas, could be other sources of this DNA, which was preserved in the clay soils.
The imprint on our own DNA
In other cases, researchers can gain insight into human history without resorting to the vestiges of the past, but through their imprint on our own DNA. Projects such as Genographic, launched by the National Geographic Society in 2005, were pioneers in mapping ancient human migrations through the similarities and differences between the genomes of today’s populations. These studies have also made it possible to determine the traces left by hybridisation with Neanderthals in modern humans of European or Asian origin, or with Denisovans in other ethnic groups. Thus, we now know that the Denisovans hybridised with modern humans, leaving some of their DNA in ours, up to 5% in the case of Australian Aborigines, Melanesians and other ethnic groups in Oceania. But we also know that up to 17% of the Denisovan genome comes from Neanderthals; in 2018, archaeologists discovered a bone fragment from a girl, given the name Denny, who lived 90,000 years ago and was the daughter of a Neanderthal mother and a Denisovan father. DNA studies have revealed that multiple interbreeding events occurred between modern humans, Neanderthals and Denisovans during the period of coexistence.
To complete the picture, scientists are even beginning to discover the imprint in our DNA of still unknown ancient species. When a team led by New York State University in Buffalo sequenced the salivary mucin protein gene 7 (MUC7) in more than 2,500 people, it was met with a huge surprise—a version of the gene present in a population of sub-Saharan Africans is distinct from all known ones, even more different than those of Neanderthals and Denisovans. The researchers concluded that the gene comes from the hybridization of Homo sapiens with another “ghost species.”
The co-director of the study, Omer Gokcumen, tells OpenMind that it is not the first time that a “gene flow from unidentified human-related species” has been found in current populations. Other studies have found traces of unknown archaic humans in our genome, and 1% of the Denisovan genome comes from an unknown ancestor, 15% of which has come down to us. “I think it is now clear that such gene flow events between hominin species, including humans, were common,” Gokcumen says. The researcher is not yet willing to wager on the identity of his ghost species, but suggests that the key will be found in the future study of ancient African DNA.
The problem, Gokcumen points out, is that climatic conditions in Africa do not facilitate the conservation of viable DNA. “The data from ancient genomes from the colder Eurasia (colder and as such the specimens are better preserved) has really changed the field,” says the geneticist. “It has been pretty difficult for African specimens,” he adds. In Europe, the oldest sequenced DNA from a modern human was extracted from 45,000-year-old bones found in caves in Bulgaria and the Czech Republic. In Africa, where our species has a much longer history, so far the oldest DNA from Homo sapiens that has been recovered is only 18,000 years old.
However, the technical barriers already overcome by paleogenetics would have seemed insurmountable only a few years ago. Experts like Rosas say that this field still holds great surprises. “We are at the beginning,” he says. “The barriers are coming down as the technique becomes more sophisticated. Ten years ago, getting 400,000-year-ago DNA would have seemed impossible.”
Comments on this publication