When Scottish physician Alexander Fleming discovered penicillin in 1928, a new era dawned for medicine that seemed to have eliminated the phantom of bacterial diseases for good. However, in 1945, when receiving the Nobel Prize, Fleming himself warned that the overuse of antibiotics could stimulate the proliferation of resistant microbes, desensitised to the effect of these drugs.
Three quarters of a century later, that prediction has become a worrying reality: in 2022, the journal The Lancet published the results of the GRAM or Global Research on Antimicrobial Resistance study, a project led by Oxford University that is the most in-depth and comprehensive to date on this “neglected pandemic”, in the words of a commentary accompanying the study.
According to its authors, a total of 4.95 million deaths worldwide in 2019 were associated with resistant bacteria, which were the direct cause in 1.27 million cases. The data make this cause of death the third leading cause of death worldwide when all associated deaths are considered, behind only heart attacks and strokes, and the 12th leading cause of direct death, trailing only COVID-19 and tuberculosis among infectious causes. The culprits of this threat are often common strains of bacteria that develop resistance to multiple antibiotics, and are popularly known as superbugs. Of the 23 included in the GRAM, only one, Streptococcus pneumoniae, has a vaccine. Here we review some of the most dangerous ones.
Resistant Escherichia coli
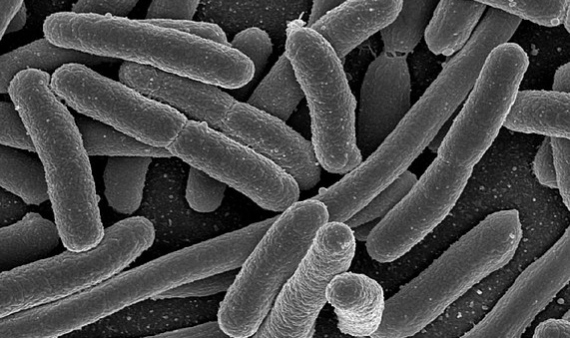
The bacterium most widely used as a tool in every biology lab in the world, and a common inhabitant of our gut flora, it is also the one that causes the most deaths due to antibiotic resistance in its most aggressive versions. According to GRAM, in 2019 Escherichia coli was associated with more than 800,000 deaths, some 200,000 directly caused by the resistant pathogen, mostly in developed countries. In addition to the ease of spread of this bacterium via the faecal-oral route, there is a particular diversity of mechanisms by which it acquires resistance to antibiotics. Of particular concern are E. coli strains resistant to fluoroquinolone antibiotics and third generation cephalosporins.
MRSA
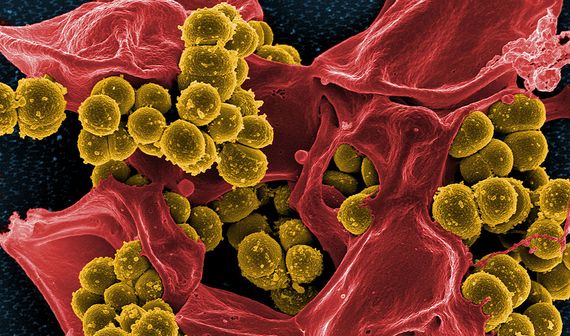
Methicillin-resistant Staphylococcus aureus (MRSA) bacterium was one of the earliest superbugs to be recognised, first detected in the early 1960s in the UK. Methicillin was at that time used to treat resistant infections, and the name MRSA began to encompass several strains of S. aureus—a bacterium common in the human microbial flora that normally only causes minor infections—that did not respond to several of the most commonly used beta-lactam antibiotics. MRSA have since become a common source of serious hospital-acquired infections, and are also very common in the general population. The resistance of S. aureus to methicillin is the pathogen-antibiotic pairing that caused the most deaths in 2019, more than 100,000, according to GRAM.
Clostridium difficile
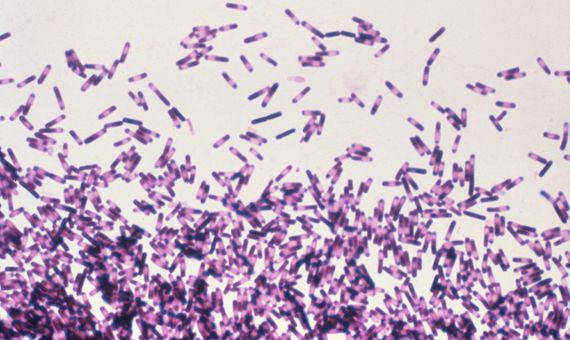
A particular case in point on this list is Clostridium difficile. While most superbugs are variants that have acquired resistance to antibiotics, this is a species that lives up to its name because it is so difficult to treat and eradicate. C. difficile is present in the gut flora of a small percentage of the population. It does not usually cause problems, but antibiotic treatment may encourage its proliferation by eliminating its weaker competitors. In these cases it can cause severe diarrhoea, intestinal damage and sepsis, especially in elderly people. The bacterium is transmitted by faecal contamination through its spores, which are resistant to alcohol disinfection.
Bacteria resistant to carbapenems

In 2017, the World Health Organization (WHO) published a document listing the superbugs for which the development of new antibiotics is most urgent. The category designated as being of critical priority brought together three groups of species that were different from each other, but with two common features. First, Acinetobacter baumannii, Pseudomonas aeruginosa and the Enterobacteriaceae family (which includes the E. coli mentioned above, along with others such as Klebsiella, Serratia and Proteus) are all Gram-negative bacteria, so called because of the presence of a double cell membrane that does not stain with the so-called Gram stain and hinders the action of antibiotics.
Secondly, these three groups include variants resistant to carbapenems, a class of potent antibiotics that are often held in reserve as a last resort when all others have failed. In 2017, the US Center for Disease Control (CDC) reported the detection in the US of more than 200 strains of these microbes, which the agency itself describes as “nightmare bacteria”. According to GRAM, more than 50,000 deaths in 2019 were due to carbapenem-resistant A. baumannii, and the same number to carbapenem-resistant Klebsiella pneumoniae. In recent years, carbapenem-resistant strains of K. pneumoniae have been described that also resist colistin, an antibiotic used only in desperate cases because of its toxicity to the kidney.
VRE

Enterococci, such as Enterococcus faecium, are another example of bacteria that live with us as part of our normal digestive flora without causing harm, but which can sometimes invade the bloodstream and colonise other tissues, causing serious infections. The 1980s saw the appearance of the first strains resistant to vancomycin (VRE), an antibiotic given intravenously to combat complicated infections such as those caused by MRSA. VREs are in the high priority category for the development of new antibiotics on the WHO list. In 2018, a study found that many circulating strains of VRE are also acquiring resistance to the alcohol used in hand sanitisers. This observation has become even more important in times of the COVID-19 pandemic, as studies have shown that overuse of disinfectant products can also promote resistance to these products and stimulate the exchange of DNA between bacteria that spreads the genes of antibiotic resistance.
Resistant gonorrhoea

Gonorrhoea, a sexually transmitted disease caused by the bacterium Neisseria gonorrhoeae, has been effectively treated with antibiotics for decades, but recently resistant strains that are difficult to treat are emerging, leading the WHO to place it in the high priority category. While diseases like this may sound like a blight of the past, the WHO counted 82 million such cases in 2020, and studies have been warning that the strains responsible are increasingly resistant to all common antibiotics, but variants resistant to even last-resort antibiotics have also been detected. The European Centre for Disease Control has warned about the risk of dissemination of these strains, which can be difficult to monitor, as many gonorrhoea infections are asymptomatic.
Resistant tuberculosis

Curiously, the WHO’s list of superbugs omits tuberculosis, a disease that in developed countries is usually associated with bygone times, but which the WHO lists as the 13th leading cause of death in the world and the second leading infectious disease after COVID-19, with more than 10 million cases and 1.5 million deaths in 2020. The reason for this omission, criticised by some experts, is not a lack of concern for this infection, but quite the opposite: previously, it had already been designated as a top priority in the search for new antibiotics. Specifically, multidrug-resistant strains of Mycobacterium tuberculosis (MDR-TB) are now a global health threat that in 2019 affected half a million people and caused 182,000 deaths, according to the Global Fund. GRAM also lists it as one of the six resistant bacteria responsible for the most deaths in 2019. The disease is especially serious in patients with weakened defences, such as human immunodeficiency virus (HIV) carriers.
Multi-resistant Staphylococcus epidermidis

A superbug that has begun to raise alarm in recent years is another species of staphylococcus, S. epidermidis, also a common inhabitant of healthy human skin. Although this bacterium is generally harmless, it can cause serious infections in immunocompromised people, a risk that has been increased by the recent detection of strains that evade the action of most common drugs, but also show increasing resistance to last-resort antibiotics such as vancomycin. The presence of the bacteria on the skin makes it difficult to avoid hospital contaminations, the most common source of these infections, and S. epidermidis has the ability to form antibiotic-resistant biofilms on plastic surfaces, including medical devices such as catheters and prostheses.
DRSP
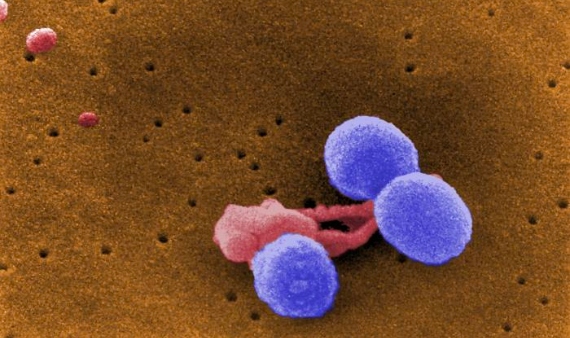
Pneumococcus, Streptococcus pneumoniae, is also a normal inhabitant of the human microbiota, in this case of the upper respiratory tract. However, it can cause serious infections in young children, the elderly or immunocompromised persons, ranging from pneumonia if it colonises the lungs to meningitis or sepsis. Antibiotic-resistant strains of S. pneumoniae (DRSP) have been known about since 1967 and have been growing at an alarming rate. Data from the US CDC indicate that more than 30% of pneumococcal infections are resistant to one or more antibiotics. These superbugs are also on the GRAM list of the six most lethal resistant pathogens in 2019. This is despite the fact that it is the only bacterium in this group for which vaccines exist. However, the WHO categorises pneumococcus as a medium priority for the development of new antibiotics, as new resistant strains emerge that escape vaccines.
Javier Yanes
Editor’s note: Original text of September 2015 updated by the author
Comments on this publication